![]()
Before you choose reagents, check that they will work with the microscope. Please ask microscopy staff. Some of the web pages here may be used as reference, such as the confocal web pages which list the available laser lines. The laser scanning confocals allow for fine tuning of spectral response for detection, but other microscope have fixed filters. This web page discusses these points. Please read below and consult with staff before choosing fluorescent probes.
There are two main things to consider when choosing fluorophores: excitation spectra and emission spectra. These are the color of light used to excite the fluorescent molecule and the color of light that you will image. (The difference between them is the Stokes shift.) The microscope you will use needs to have both the correct excitation and the correct emission.
The filter blocks on most fluorescent scopes are configured for the emissions of blue, green, red, near infra-red as pictured. This was standard. By special request, other filter blocks could be added. And a few of the confocals could also handle CFP and YFP similar fluorescnt proteins.
These curves show the emission characteristics of the standard configuration on the Zeiss AxioObserver and most other fixed filter microscopes.
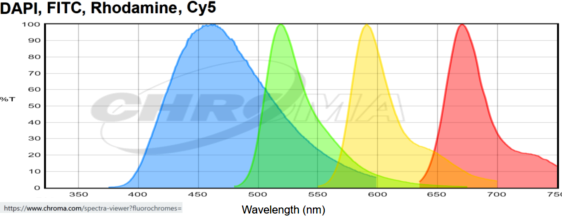
In 2023, two microscopes with farther red detection as a standard feature were added. This graph is an example of fluorescent probes that could be used in a single sample. This combination is of Dapi, AF488, AF568, AF647, and AF750 with emission curves generated at https://www.chroma.com/spectra-viewer?fluorochromes=800%2C47%2C48%2C53%2C57%2C163

The two scopes which can image the additional far-red probes are the Crest X3 spinning disk confocal and the Leica Stellaris laser scanning confocal. Check each web page for deatils of excitation wavelengths and available emission ranges.
There is an additional way to add probes which is to distinguish them by excitation wavelength. Probes with the same emission spectra may be imaged in sequence. This email exchange explains the strategy.
The curves above do not show excitation spectra. In general, for a widefield microscope such as the Zeiss AxioObserver:
| ex (nm) | em (nm) | |
| blue | 365 | 460 +/- 40 |
| green | 490 | 525 +/-25 |
| red | 560 | 590 +/- 25 |
| near IR | 640 | 650 long pass |
Note that the standard Dapi/Hoechst filter set will not work with fluorophores that excite at 405 nm. For instance, Alexa 405 and Pacific Blue are bad choices for widefield scopes with standard Dapi/Hoechst filter sets that excite at 365 nm. However, the 405 nm laser on most confocal microscopes work efficiently with these probes and with the Brilliant Violet probes. Also, whereas Dapi works well with the 405 laser, Hoechst does not.
Therefore, you need to choose the microscope before selecting which blue probe you will use.
For CFP and mTurquoise type fluorescent proteins, see below.
The Zeiss 880 has a 34 channel array that can be used for spectral deconvolution. For example, with a panel of Opal fluorochromes used for the Vectra.
Note that Alexa 594 does not neatly fit these standard filter sets. For some reason, Alexa 594 has become incredibly popular even though it doesn't really fit any of the standard fluorescent microscope filter sets. Using a standard red filter, you can see a signal, but not efficiently. But the confocal microscopes with 594 nm lasers work extremely well with this fluorophore except that it may spill into a longer red channel and slows down image collection do to moving parts in the microscope. In general, we tell people to use Alexa 545, 555, or 568 (or similar spectra to Cy3, Atto 550, DyLight 549, TRITC) instead and then skip to the next channel with Alexa 633 or 647. (On the other hand, onthe Zeiss 880 or Leica SP8 you may be able to sandwich it between a bright red and a bright near infra-red signal, but we only recommend this when identifying different cells types that only stain with one probe to avoid spillover that would make analysis ambiguous.)
Probes out more in the IR are becoming more popular. However, standard laser scanning confocal microscopes do not detect light longer than 750 nm and are also limited by the discrete laser lines for excitation. Looking at the spectra for Alexa 750 and Cy7, for instance, it excites inefficiently between 630 and 647 nm, which is the range of lasers used on most confocals, and emits above 750 nm, which is outside the detection range. But CCD and sCMOS cameras detect into the IR very well and microscopes with bright white lamps (metal halide) may access the wavelengths needed for exciting the longer wavelength probes. By special request, a Cy7/Alexa750 filter block is available for the Zeiss AxioObserver microscope. However, due to chromatic aberration, it involves changing the focus when being used with other fluorphores. And as discussed below, there are two scopes in the core which can routinely image out to 850 nm.
Using online tools for fluorescent spectra, both excitation and emission, and with detailed knowledge of the available microscopes, we can help you tailor your panel of probes to match the correct microscope.
IMPORTANT: When using multiple antibodies, there is an increased chance of cross-reactivity. Also, antifbodies or other labeling methods may compete for binding sites. Therefore, design robust controls.
Excellent resources for probes:
https://www.janelia.org/open-science/jfx-dyes
https://biotium.com/technology/cf-dyes/
For TEM? https://www.cell.com/cell-chemical-biology/pdf/S2451-9456(20)30243-9.pdf
The following tables are guidelines of probes that match the dominant filter sets configurations on our microscopes. They is not complete. They are meant to provide a guide to selecting fluorescence probes.
For AxioObserver widefield:
| Blue ex 365 nm | Green ex 488 nm |
Red ex 565 nm |
Near Infra-Red ex 640 nm |
| Dapi, Hoechst | AF488, Atto 488, DyLight488, eGFP, FITC, Lucifer yellow | Cy3, AF555, AF568, CF555, CF568, Atto 550, RFP, mCherry, DyLight 549 | Cy5, AF633, AF647, Atto 633, DyLight633 |
As stated above, there are some variations for the confocals, so please contact us with any questions.
One question we often get is, what is the brightest fluorescent protein. We have been relying on FPbase. For example, here are comparisons of a few typical green fluorescent proteins that all mature in 10 to 15 minutes.
| ex | em | QY | brightness | |
| mNeonGreen | 506 | 517 | 0.8 | 92.8 |
| sfGFP | 485 | 510 | 0.65 | 54.2 |
| mGreenLantern | 503 | 514 | 0.72 | 73.3 |
| EGFP | 488 | 507 | 0.6 | 33.5 |
| Emerald | 487 | 509 | 0.68 | 39.1 |
Microscopes that can image CFP and mTurquoise type probes are:
Sent: Friday, January 5, 2024 10:40 AM Ideal ex for mTurquoise2 is 435 nm. We have three microscopes with lasers at or very near this wavelength. We also have filter blocks which can be swapped into these scopes on request: And, of course, the 2P scope can excite and detect this probe too, although sometimes post-processing is required to separate from the green channel. This can be done robustly as demonstrated in https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3461908/ |
===============================================================================
Mitochondria probes recommended on the confocal listserv in early 2024:
Abberior Live Orange, an internal cristae dye, cationic probe reliant on membrane potential
BacMam CellLight Mitochondria probes, commercially available from Thermo Scientific, target E1 alpha pyruvate dehydrogenase, are independent of membrane potential, and retain decent signal post PFA fixation for standard IF studies.
This paper https://www.nature.com/articles/s41589-023-01450-y compares MAO-SiR and mEmerald-TOMM20 and other variations.
===============================================================================
To: Cammer, Michael <Michael.Cammer@nyulangone.org>
Subject: Re: Can I add 5th marker in my staining Panel
I have
Fascin - AF546
Tcrb - AF488
MHCII- BV421
FoxP3 - AF647
From: Cammer, Michael
Subject: Re: Can I add 5th marker in my staining Panel
There are a few strategies. I am not going to list all, but here are the simplest.
1. The BV dyes that excite at 405 nm can be used with the LSM880 you use now. https://www.thermofisher.com/us/en/home/life-science/cell-analysis/flow-cytometry/brilliant-polymer-dyes.html
Based on the brightness scale, BV605 looks like the best candidate. BV786 will not work. Make sure BV labeled features are not in the same locations as other features with the same emission range.
2. Switch to the new spinning disk confocal and add a probe that excites at 730 nm. https://microscopynotes.com/crest01/index.html
3. Switch to the Leica Stellaris and use any probe with an excitation between 690 and 790 nm with an emission less than 850 nm. https://microscopynotes.com/stellaris/index.html
If there are any overlaps in labeling, then we would need controls to check for cross reactivity.
===============================================================================
Need to uniformly label structures in cells, such as nuclei? Here is an example:
"We used the following iPSC cell line for all experiments (also see Supplementary methods table 1) : Histone2B-mEGFP that uniformly labels nuclei (cell line ID: AICS-0061-036), mEGFP-Beta-Actin that uniformly labels ACTB (cell Line ID: AICS-0016 cl.184), mTagRFP–T–CAAX which labels cell membrane (cell line ID: AICS-0054-091), mTagRFP–T-Tubulin-alpha1b that labels TUBA1B (cell line ID: AICS-0031-035), mTagRFP–T-LaminB1 that labels LMNB1 (cell line ID: AICS-0034-062) and unlabeled WTC iPSCs (cell line ID GM25256)." https://www.biorxiv.org/content/10.1101/2023.08.21.553827v1.full
[need to link to quenching webpage too... and discuss controls at same settings.]