 Zeiss 700 microscope
Zeiss 700 microscope
Alexandria Center East 3rd floor, 349 (click here for a map)
Badge Access Request Here
Phone in room x42677 or 646-754-2677
To use microscope you must be trained by Michael Cammer or Yan Deng via the Microscopy Core.
** Sign up to use microscope ** Official Zeiss instructions "Quick Guide"
Startup:
- Appointment in iLab.
- Only if need environmental chamber heat on, power strip #3 for incubator.
- Hg arc lamp for viewing fluorescence by eye.
- Power strip switch #1.
- Power strip switch #2.
- Computer:
- Check if already on. If logged in, then log out and log in.
- If off, turn on with power on front of computer.
- Zen software. Do not touch microscope while blue bar is progressing during software startup.
- If you get a Keyserver error, this means that you must start a session using ilab Kiosk.
- Make sure the microscope stage plate is properly inserted flat and recessed. Before doing this, make sure the lens is at the lowest focus position.
- Save all files in CZI format.
- Save all files in Drive D:
Shutdown:
- If oil lens used, lightly clean.
- Switch to 10X lens.
- Focus the microscope as low as it will go. The touch pad will Say Lowest Z Limit.
- Copy your files over network or to USB device to take with you.
- Quit software.
- Log out of computer account. Computer may be left on.
- Switches in reverse order.
- DO NOT turn off computer monitor
|
Please read this document: https://microscopynotes.com/700/safety_update_20230411.pdf
General Confocal Best practices:
- The pinhole is what makes the confocal a confocal. 1AU (which means 1 Airy unit) or smaller (0.5 to 1.0 ok) and click the 1AU button each time you change lenses.
If you are opening it for imaging fixed samples, you should go use a widefield fluorescence scope instead.
Except in special case of live cell imaging where you understand that images are not confocal, this is NOT AN ACCEPTABLE WAY TO MAKE IMAGES BRIGHTER. You won't hurt the instrument, but when you write your methods, you won't be accurately describing your microscopy as "confocal".
- Offset. Always use at 0 or 1 (or greater than one in some rare cases).
Negative numbers are wrong.
- Digital gain. The preset is 1. Leave it there.
- Zoom 1 or greater. Using a Zoom less than 1 may cause a hot spot &/or curvature at the edges.
- Avoid saturation. Saturating images 1.) makes features appear larger than they are, 2.) prevents quantification of intensities and 3.) may damage the detectors inthe microscope.
Use the Range Indicator button to make sure you have no saturated pixels. If you see red pixels, you need to turn down the Gain or Laser.
Turning gain down will reduce noise. Less noise means you can scan faster. (More here.)
- Always save images in .czi format. This preserves each channel and all instrument metadata.
Why take images at high resolution of a confocal and then throw away this information by saving as lossy jpg?
Highly recommended reading: Tutorial: guidance for quantitative confocal microscopy
About the microscope
| Laser lines |
Useful for... |
| 405 nm |
Dapi, CFP, Pacific Blue, Alexa 405, Brilliant Violets |
| 488 nm |
FITC, Alexa488, GFP, YFP, NBD |
| 555 nm |
RFP, mCherry, rhodamine, Cy3, Alexa568, Alexa594, propidium iodide |
| 639 nm |
Cy5, Alexa647, To-Pro |
Lenses:
- 10X N.A. 0.45 air
- 20X plan apochromat N.A. 0.75 air DO NOT GET OIL ON THIS LENS!
- 40X plan apochromat N.A. 1.4 oil
- 63X plan apochromat N.A. 1.4 oil
- 100X plan apochromat N.A. 1.4 oil (recommend do not use)
Setting up multiple channels for detection:
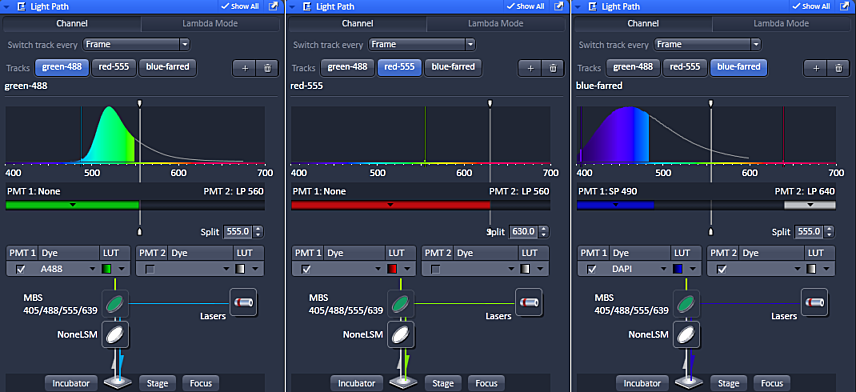
There are two ways to choose the spectral detection. One is by filter and the other is by a user adjustable mask. If you are using multiple channels, imaging will be much faster if the mask does not move between channels. We typically leave it at 555 nm. But custom selection may provide fine tuning of spectral detection.
In the above example, the splitter is moved for the red channel because there is no short pass 630 filter in the system to allow for red only collection. If the splitter were not set at 630 nm with the red detection in PMT1, then there is a possibility that far-red probe excited by the 555 nm laser could be detected as spill over in the red channel.
There is no worry of green being imaged in this channel because fluorescence is always red shifted; a photon of less energy is emitted.
Method for Manual uncaging / photo activation using widefield illumination.
Partial list of publications with data from this microscope, last updated 2018:
- Azoulay-Alfaguter I, Strazza M, Pedoeem A, Mor A. The coreceptor programmed death 1 inhibits T-cell adhesion by regulating Rap1. J Allergy Clin Immunol. 2015 Feb;135(2):564-7. doi: 10.1016/j.jaci.2014.07.055. PMID: 25240786
- Azoulay-Alfaguter I, Mor A. Proteomic analysis of human T cell-derived exosomes reveals differential RAS/MAPK signaling. Eur J Immunol. 2018 Sep 12. doi: 10.1002/eji.201847655. PMID: 30207595
- Chenette, DM, Cadwallader, AB, Antwine, TL, Larkin, LC, Wang, J, Olwin, BB, Schneider, RJ. Targeted mRNA Decay by RNA Binding Protein AUF1 Regulates Adult Muscle Stem Cell Fate, Promoting Skeletal Muscle Integrity. Cell Reports. DOI: http://dx.doi.org/10.1016/j.celrep.2016.06.095
- Daley D, Mani VR, Mohan N, Akkad N, Pandian GSDB, Savadkar S, Lee KB, Torres-Hernandez A, Aykut B, Diskin B, Wang W, Farooq MS, Mahmud AI, Werba G, Morales EJ, Lall S, Wadowski BJ, Rubin AG, Berman ME, Narayanan R, Hundeyin M, Miller G. NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. J Exp Med. 2017 Apr 25. pii: jem.20161707. doi: 10.1084/jem.20161707. PMID: 28442553
- Dann R, Hadi T, Montenont E, Boytard L, Alebrahim D, Feinstein J, Allen N, Simon R, Barone K, Uryu K, Guo Y, Rockman C, Ramkhelawon B, Berger JS. Platelet-Derived MRP-14 Induces Monocyte Activation in Patients With Symptomatic Peripheral Artery Disease. J Am Coll Cardiol. 2018 Jan 2;71(1):53-65. doi: 10.1016/j.jacc.2017.10.072. PMID: 29301628
- Greco SH, Torres-Hernandez A, Kalabin A, Whiteman C, Rokosh R, Ravirala S, Ochi A, Gutierrez J, Salyana MA, Mani VR, Nagaraj SV, Deutsch M, Seifert L, Daley D, Barilla R, Hundeyin M, Nikifrov Y, Tejada K, Gelb BE, Katz SC, Miller G. Mincle Signaling Promotes Con A Hepatitis. J Immunol. 2016 Aug 24. pii: 1600598. PMID: 27559045
- Peled M, Dragovich MA, Adam K, Strazza M, Tocheva AS, Vega IE, Mor A. EF Hand Domain Family Member D2 Is Required for T Cell Cytotoxicity. J Immunol. 2018 Oct 1. pii: ji1800839. doi: 10.4049/jimmunol.1800839. PMID: 30275048
- Peled M, Tocheva AS, Sandigursky S, Nayak S, Philips EA, Nichols KE, Strazza M, Azoulay-Alfaguter I, Askenazi M, Neel BG, Pelzek AJ, Ueberheide B, Mor A. Affinity purification mass spectrometry analysis of PD-1 uncovers SAP as a new checkpoint inhibitor. Proc Natl Acad Sci U S A. 2017 Dec 27. pii: 201710437. doi: 10.1073/pnas.1710437115. PMID: 29282323
- Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, Avanzi A, Barilla R, Daley D, Greco SH, Torres-Hernandez A, Pergamo M, Ochi A, Zambirinis CP, Pansari M, Rendon M, Tippens D, Hundeyin M, Mani VR, Hajdu C, Engle D, Miller G. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016 Apr 14;532(7598):245-9. doi: 10.1038/nature17403. PMID: 27049944
- Strazza M, Azoulay-Alfaguter I, Dun B, Baquero-Buitrago J, Mor A. CD28 inhibits T cell adhesion by recruiting CAPRI to the plasma membrane. J Immunol. 2015 Mar 15;194(6):2871-7. doi: 10.4049/jimmunol.1401492. PMID: 25637021
- Su W, Wynne J, Pinheiro EM, Strazza M, Mor A, Montenont E, Berger J, Paul DS, Bergmeier W, Gertler FB, Philips MR. Rap1 and its effector RIAM are required for lymphocyte trafficking. Blood. 2015 Dec 17;126(25):2695-703. doi: 10.1182/blood-2015-05-644104. PMID: 26324702
- Wang W, Marinis JM, Beal AM, Savadkar S, Wu Y, Khan M, Taunk PS, Wu N, Su W, Wu J, Ahsan A, Kurz E, Chen T, Yaboh I, Li F, Gutierrez J, Diskin B, Hundeyin M, Reilly M, Lich JD, Harris PA, Mahajan MK, Thorpe JH, Nassau P, Mosley JE, Leinwand J, Kochen Rossi JA, Mishra A, Aykut B, Glacken M, Ochi A, Verma N, Kim JI, Vasudevaraja V, Adeegbe D, Almonte C, Bagdatlioglu E, Cohen DJ, Wong KK, Bertin J, Miller G. RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer. Cancer Cell. 2018 Nov 12;34(5):757-774.e7. doi: 10.1016/j.ccell.2018.10.006. PMID: 30423296
LSM 700 Serial# 2601000862 which is currently located in Alexandria East
<-- Back
comments, questions, suggestions for this web page: Michael.Cammer@med.nyu.edu or mcammer@gmail.com
 Zeiss 700 microscope
Zeiss 700 microscope Zeiss 700 microscope
Zeiss 700 microscope