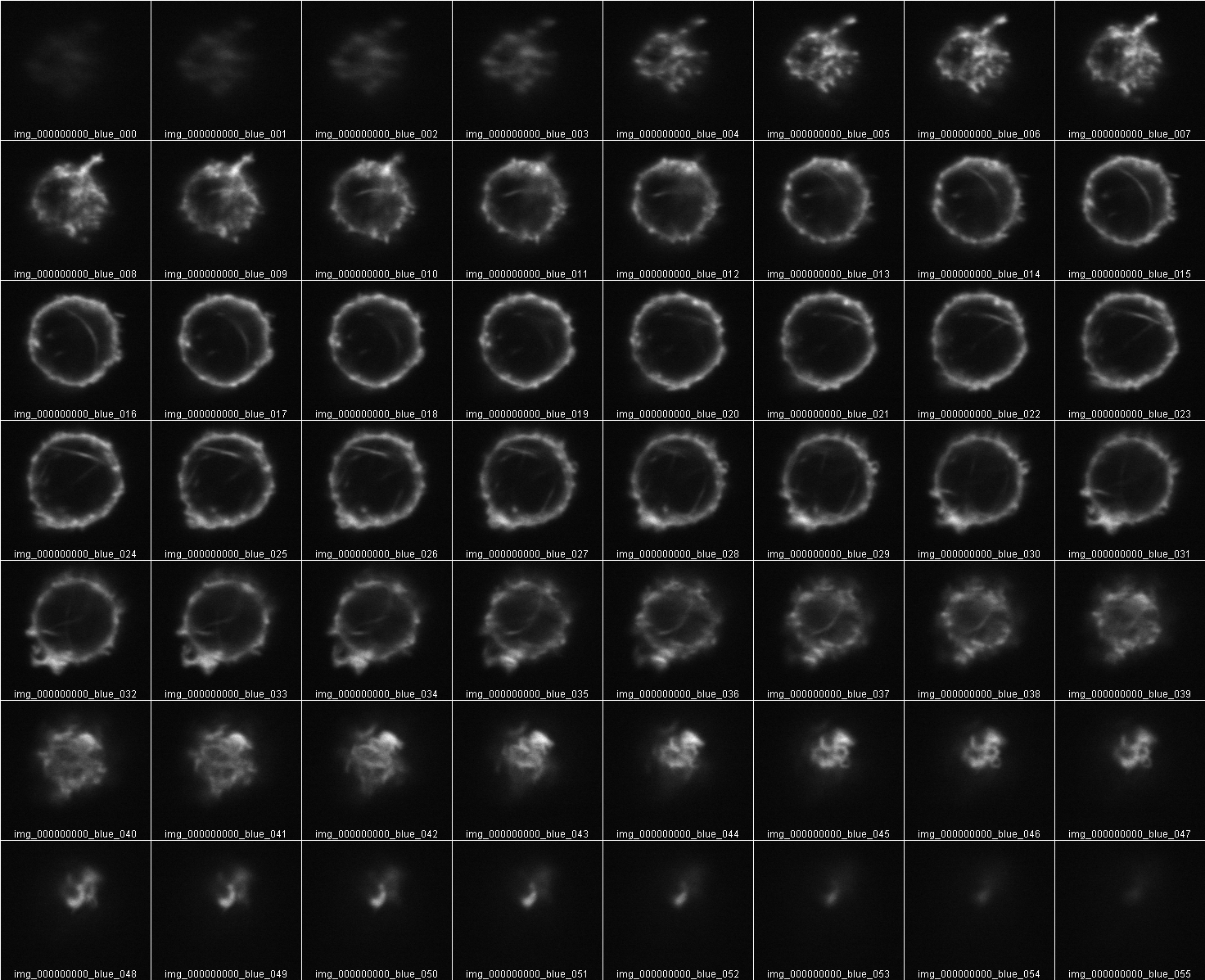
Measure Intensity of a Z stack
"I need to quantify fluorescence intensity for a whole cell, but do not know how to do this. Once I create a Z stack of the cell, how do I go about telling Image J how to identify the cell and then to quantify the fluorescence intensity?"
Before discussing ImageJ we should discuss measuring total intensity (as a metric for total amount of a molecular species) in individual cells.
A low numerical aperture lens with widefield fluorescence can capture the entire depth of a cell in one shot (if you don’t have a widefield scope, you can open the pinhole on the confocal, although the PMT is not as good a detector for this as a camera). If you are looking for whole cell measurement, then it is simple to measure each cell once to get the answer. Also, this means you can image a far greater number of cells faster. You may even want to look to see if anyone there has a plate reader that takes images rather than just one intensity reading for the whole field; this can make the analysis almost high throughput.
Confocal Z stacks at high magnification are really only needed if you want to measure the specific compartment within the cell where the fluorescence is located or if cells are piled up on top of each other.
To measure total fluorescence from a Z series the sectioning must begin in the dark and end in the dark. In other words, the entire volume must be included. No top or bottom slices cut off.

But this Z stack also is oversampled. It will give twice the sum of intensities as the less often sampled in Z volume:
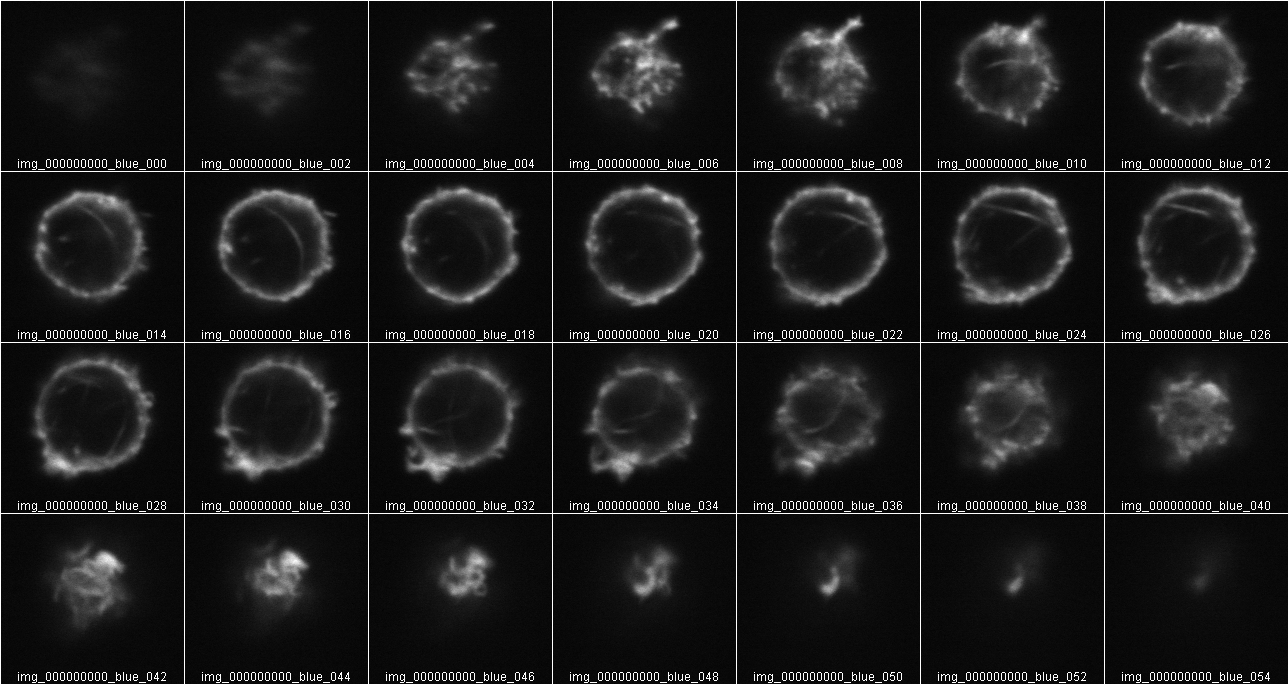
This points out that all original data must be collected at the same X,Y zoom and Z step size. What should the Z step size be? Don’t waste time oversampling. But don't undersample too much.
Essentially, to get the total intensity of each cell you can isolate the cell from its neighbors and then step through to measure the intensity at each Z section. But before doing that, let's subtract the background; this will eliminate unlabeled voxels from the counted volume.
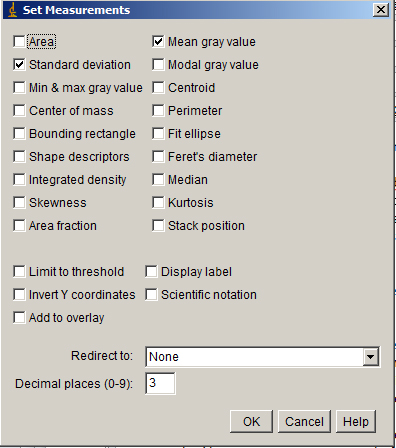
Analyze > Set Measurements... will give you a menu. Check Mean and Standard Deviation.
Measure a background value around the cell to subtract from the entire volume. (You can also subtract the background plus the standard deviation; just be consistent.)
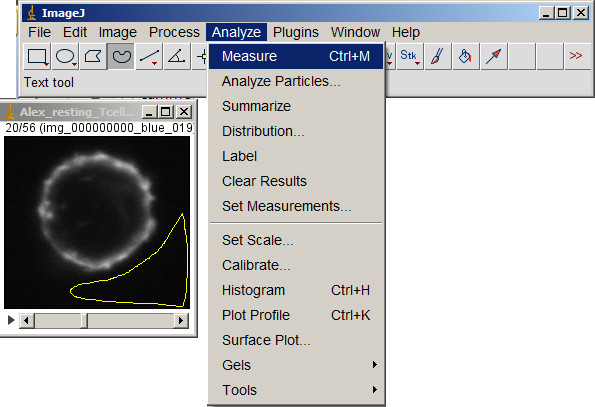
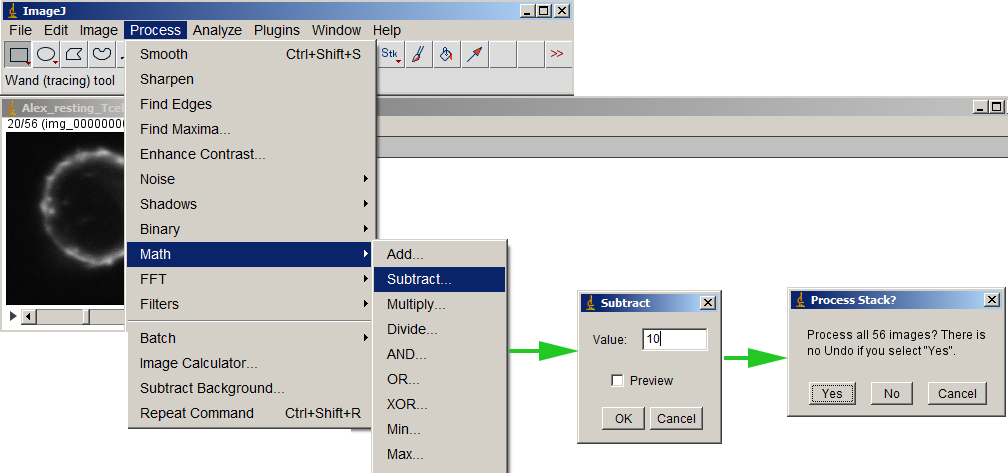
Make sure either nothing in the image is selected or Select All. Then subtract the bg value from the entire volume.
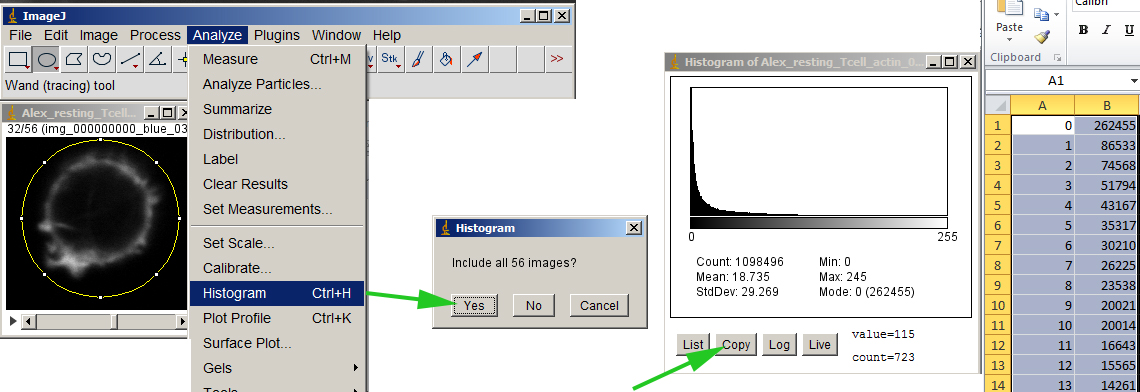
Measure the histogram. This will give you a list of the number of pixels at each intensity value. Copy and paste into your favorite spreadsheet.
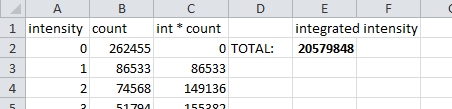
Multiply each intensity by the count of voxels at that intensity. Then sum to get the integrated intensity. See the spreadsheet.
ImageJ can also do the calculation with a brief macro.
Also, now that I wrote all this down, I realize there may be a plugin out there on the web somewhere that will do all this. Regardless, if this is something that you will have to do a lot, it could be mostly automated with a macro that is easy for me to write.
But you can see that the low NA image collection method followed by a background subtraction followed by measuring each cell is so much faster.