The idea of giant servers in the sky with all data available to anyone who can pay or hack in is not a new one. My first experience with this was in the mid-1980s reading science fiction.
No longer is this the purview of fiction. Big server farms can hold large amounts of data with varying levels of accessibility to the outside world. The data may be highly indexed or it may be a dump.
Microscopy imaging from research labs resembles a dump far more than tagged.
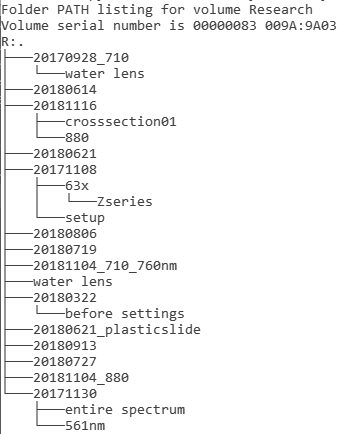
Look, for instance, at these two directory trees drawn arbitrarily from a server containing data from multiple labs using a shared confocal resource.
| Tree showing all diectories in a user's data container. Note that none of these names refer to biological or chemical conditions. |
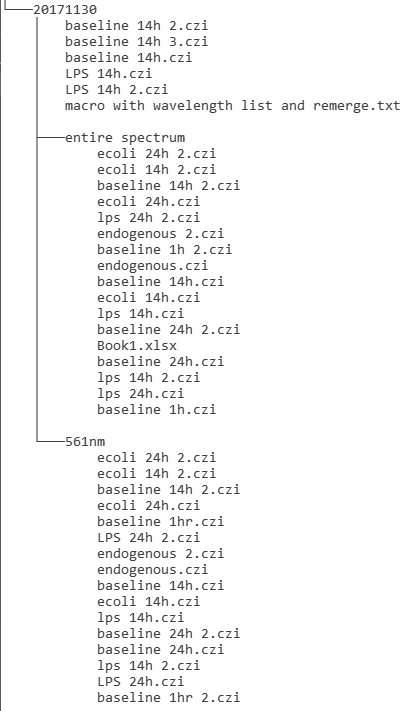
One date expanded to show how the data from the day's imaging was named. How many experiments does this repesent? What were the experiments about?
(The "czi" extension is Zeiss's format which contains extensive metadata about instrument parameters.)
|
 |
 |
In some respects, this is better than some naming. It's not uncommon to find folders labeled by date in a format such as "3-7-12" with folders inside called "control" and another labeled "ko" with files named "image1.jpg", "image2.jpg" where, clearly, the jpg files are compressed and contain no metadata at all. As I write this, from the two instruments used the server shows more than 8.3 TB of data in 200,000+ files not including the files not ever put into our shared server space.
Is "3-7-12" March seventh or July third? Or is this really December 7, 2003? Perhaps the data still retains file creation date to resolve the problem, but after multiple copies, this information may be lost.
This is how biologists all around the world are storing their data.
The dates refer to notebooks.
From 1991 through 2008 I did confocal imaging for a biologist who named each slide with a unique number. We started with a letter and three digit numbers back when file names were limited to eight characters and we retained this convention. She kept impeccable and detailed notes about the biological and preparation conditions of each slide we imaged. There was a hand written master list of the slide and file names. This notebook, however, referred to other handwritten notebooks with the extensive details. Today (2018) I have all the files on external hard drives. But where are the notebooks?
This is the rule, not the exception. Actually, maybe it is the exception as I doubt that most of the people doing imaging have such excellent notes characterizing the images at any step of the experiments.
And since then experiments have gotten far more complex. Instead of simple single field images with Z series, now we have live imaging with many parameters changing, the experiments themselves moving targets. And much of the data are failures, false starts, compromised which are not deleted from storage; they persist on taking up space.
Could these data be usefully searched?
It would require an enormous cultural shift for the data to be indexed. Every dataset could have multiple tags or pointers to other databased containing cell types, media type, etc. A person tethered to a computer would have to enter these data. The infrastructure would have to exist to make entering these data possible. And they would have to be entered properly.
Some settings could be logged automatically Let's say an experiment is supposed to be run at 37 degrees C with 5% CO2. This would need to be captured and this could be captured automatically by a microscope programmed to do so.
But the user would have to enter all the biological and chemical conditions. Is this going to happen?
PI says, "Let's see how our protein behaves in CD45 positive T cells." So the student hunts down a lab that can provide expertise in these cells. Will the entire chain of prep from the type of mouse to isolation technique to culturing conditions to media to coating on the culture dish make it into the metadata?
Tagging after the fact incomplete.
Ten years ago a PI complained that when he looked at data from the many students and postdocs in his lab, he often id not know what he was looking at. Therefore, he insisted that they put a simple text file in every directory of images and that the text file move with the images.
Name:
Date of imaging:
Reference to date in paper notebook:
Goal: (phrase or brief sentences explaining why this experiment)
Microscope being used:
Objective magnification:
Probes used and the names of the dyes: (example: rhodamine anti-beta tubulin from Sigma)
Channel 1:
Channel 2:
Channel 3:
Channel 4:
Channel 5:
Channel 6:
Channel 7:
Channel 8:
Cell types or mice strains:
What is the substrate, for instance, if bilayers, what proteins are in the bilayers at what concentration for each condition?
Fixation (chemistry, time, temp):
Other treatments:
Z step size:
Time interval:
Explain the directory and file naming conventions:
Non-compliance as uniform across the lab.
When his lab moved, he imposed an electronic notebook requirement. Unfortunately, I do not know whether imposing electronic notebooks solved this problem. Based on apparent SOP by most labs, my guess is that microscopy experiments often were not logged. But maybe this is unfair on my part. Except for seeing the file names, I really don't know what happens to the data once they are copied to individual lab data silos.
More important: this isn't a problem. This is how labs have always performed. It is defined as a problem by papers such as https://www.nature.com/articles/s41592-018-0195-8 [local copy] and cell imaging library and https://idr.openmicroscopy.org/about/ or by government calls for archiving.

